
Development of a genetic barcode for surveillance and malaria parasite tracking in Sri Lanka
Parasitic Diseases Research Unit, Department of Parasitology, Faculty of Medicine, University of Colombo in collaboration with the Broad Institute of MIT and Harvard, Cambridge, USA
Sri Lanka was certified as a malaria-free nation in 2016, but imported malaria cases continue with a persistent risk of re-emergence of the disease in the country
Genetic information of a parasite population may be used to understand the trends in transmission as well as identify imported parasites to support elimination programs. A genetic barcode already exists for P. falciparum (the other common human malaria parasite) that may be used for tracking parasites to its source. A single nucleotide polymorphism (SNP) barcode specific for Sri Lankan P. vivax parasites was developed for the first time to evaluate the population dynamics of P. vivax parasite isolates from Sri Lanka and to assess its ability for tracking the parasites of uncertain origin to its geographic location. The scientists at the Department of Parasitology, Faculty of Medicine, Colombo, collaborated with the Broad Institute of MIT and Harvard, Cambridge, USA, which is a leading institution in genetics and bioinformatics research. The barcode was developed using 51 P. vivax samples collected during 2005-2011, mainly from three provinces of the country. Forty SNPs were genotyped using a high-resolution melting (HRM), single-nucleotide barcode method.
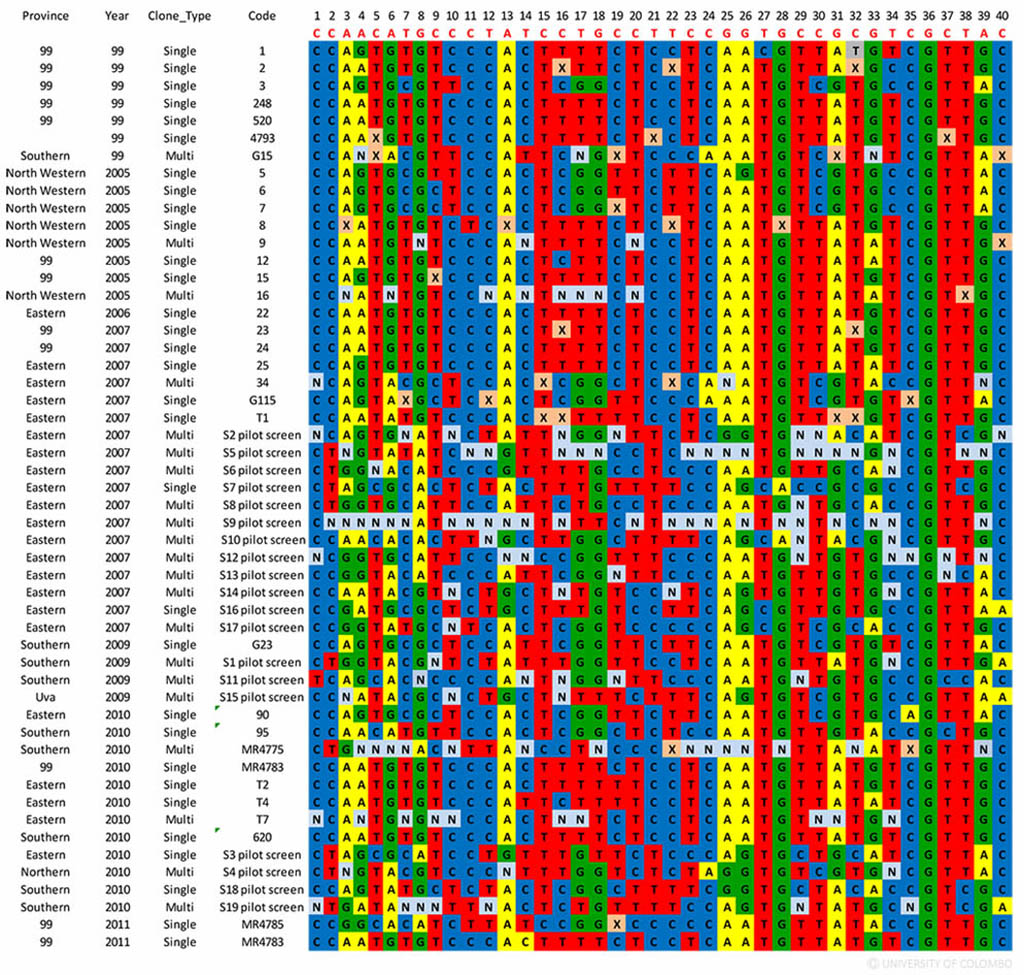
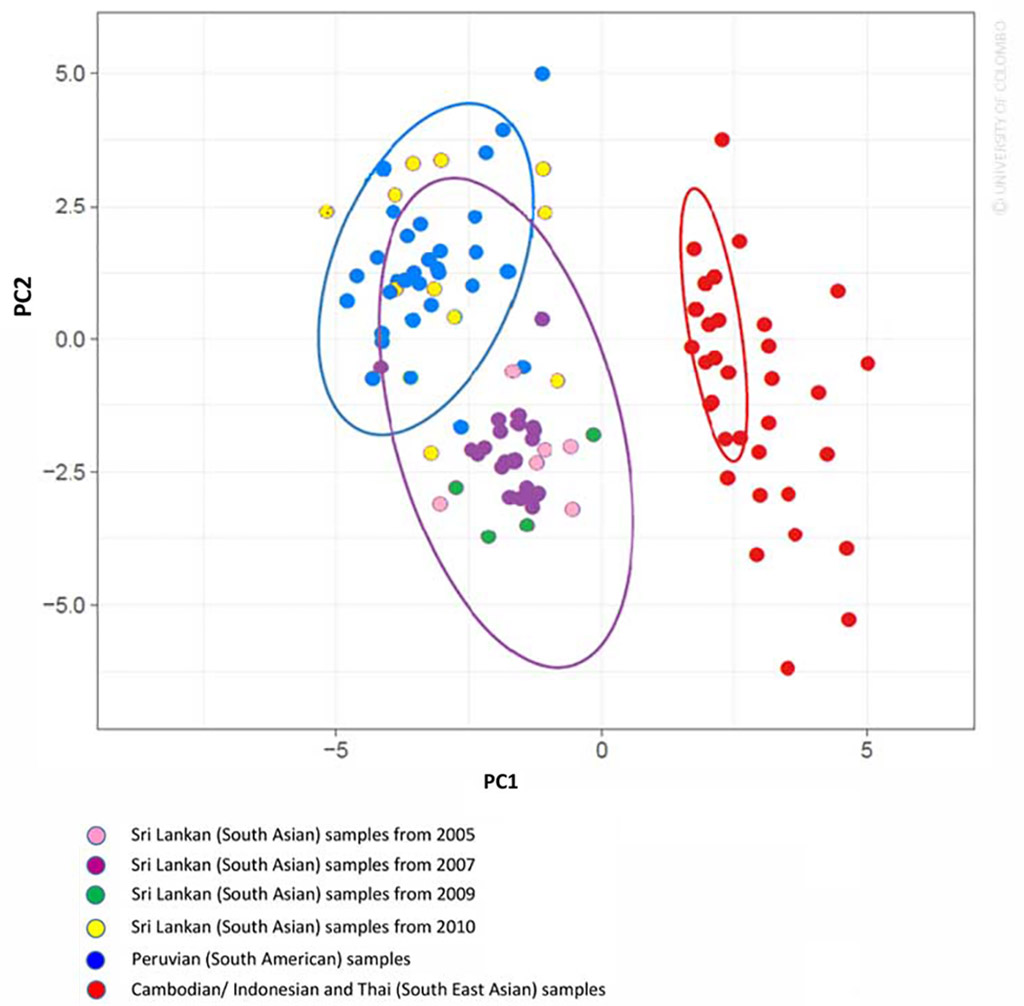
The SNP barcode for the Sri Lankan P. vivax parasites – the barcode enabled to distinguish between South American, South East Asian and South Asian parasite isolates in principal component analysis. The barcode could also be useful in assessing genetic diversity of the local parasite population.
Malaria Journal publication in September 2020: Dewasurendra et al. (2020)
https://malariajournal.biomedcentral.com/articles/10.1186/s12936-020-03386-3
THE TEAM

Prof. Nadira Karunaweera
Lead investigator

Prof. Sharmini Gunawardena
Lead investigator

Dr. Rajika Dewasurendra
Lead investigator

Mr. G. M. G. Kapilananda
Research supportive staff

Mr. M. Mohan
Research supportive staff
Department of Parasitology, Faculty of Medicine, University of Colombo

Dr. Sarah Volkman

Dr. Daniel Neafsey

Dr. Stephen Schaffner

Prof. Dyann Wirth

Dr. Mary Lynn Baniecki










